A team led by Professors Guo-Qiang Bi and Pak-Ming Lau from the Hefei National Research Center for Physical Sciences at the Microscale and the Division of Life Sciences and Medicine at the University of Science and Technology of China (USTC), in collaboration with the Institute of Artificial Intelligence, Hefei Comprehensive National Science Center (IAI) and Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences (SIAT), has made a major breakthrough in the field of three-dimensional (3D) imaging of large-scale biological tissues. They developed the world’s fastest high-definition 3D imaging technology for the entire body of small animals at subcellular resolution, enabling efficient mapping of the fine architecture of the peripheral nervous system (PNS). The findings were published in Cell on July 10th (Beijing Time).
3D Visualization of Peripheral Nerves Throughout the Mouse Body
The PNS serves as the body’s "internet of things", mediating bidirectional communication and modulation between the brain and organs: transmitting motor commands to regulate critical functions like breathing and heartbeat, while processing and relaying back sensory signals such as pain and temperature. This enables efficient physiological coordination among various tissues and organs. Mapping the intricate connections of the PNS throughout the whole body is essential for fundamentally understanding its complex functional mechanisms and related diseases pathogenesis.
For a long time, knowledge of PNS architecture relied on millimeter-resolution anatomical studies. Over the past decade, advances in 3D optical microscopy have propelled micron-resolution whole-brain mesoscopic connectomic mapping, but similar analyses for the PNS across the whole body have remained challenging. Existing imaging techniques struggle to balance imaging resolution and speed. Even when combined with whole-body sample clearing techniques, resolving the long, complex, and often intermingled nerve routes of the PNS at the whole-body scale remains challenging.
The team previously developed a volumetric imaging with synchronized on-the-fly scan and readout (VISoR) technology for 3D imaging of cleared thick-sectioned brain samples. VISoR possess the advantage of high imaging speed, high resolution, and scalability. It has achieved imaging of a whole mouse brain within 1.5 hours at sub-micron resolution (Hao Wang, Qingyuan Zhu, et al., National Science Review, 2019), and 3D imaging of a whole macaque brain and single-fiber tracing after optimization for the first time (Fang Xu, Yan Shen, Lufeng Ding, Chao-Yu Yang, et al., Nature Biotechnology, 2021). However, this section-then-image approach is not suitable for whole mouse bodies. Unlike the relatively compact and homogeneous brain, the mammalian body is much larger and highly heterogeneous, containing irregular structures and diverse tissue types. It often leads to deformation and damage of body slices prior to VISoR imaging, making complete reconstruction difficult.
To overcome these challenges, the team pioneered a strategy of " in situ sectioning + 3D blockface imaging". They developed a blockface-VISoR imaging system integrating a precision vibratome, and the ARCHmap protocol for whole-body clearing and hydrogel embedding. Each cycle captures ~600 μm-depth 3D surface images before automatically removing 400-μm-thick tissue, repeating until completion. Automated inter-section stitching algorithms then perform seamless 3D alignment using ~200-μm overlapping regions between adjacent sections. The shallow imaging depth minimizes light scattering in cleared tissue, enabling high resolution. Based on this strategy, the researchers established an optimized technical pipeline, and achieved uniform subcellular-resolution 3D imaging of an entire adult mouse body within 40 hours, generating ~70 terabytes of data per fluorescence channel. Over 4 petabytes of raw data from dozens of mice have been collected.
Due to the excellent fluorescent signal preservation of the sample processing method, the researchers verified that the technology is compatible with commonly used labeling methods in neuroscience, such as transgenic labeling, virus labeling, and immunostaining. Combined with imaging, the researchers analyzed the fine structure and single-fiber projection paths of different types of peripheral nerves throughout the mouse body. They revealed the cross-segmental projection characteristics of single spinal neurons for the first time, mapped the organ-specific vascular distribution pattern of sympathetic nerves throughout the body, and resolved the overall projection architecture of the vagus nerve and the complex single-fiber routes.
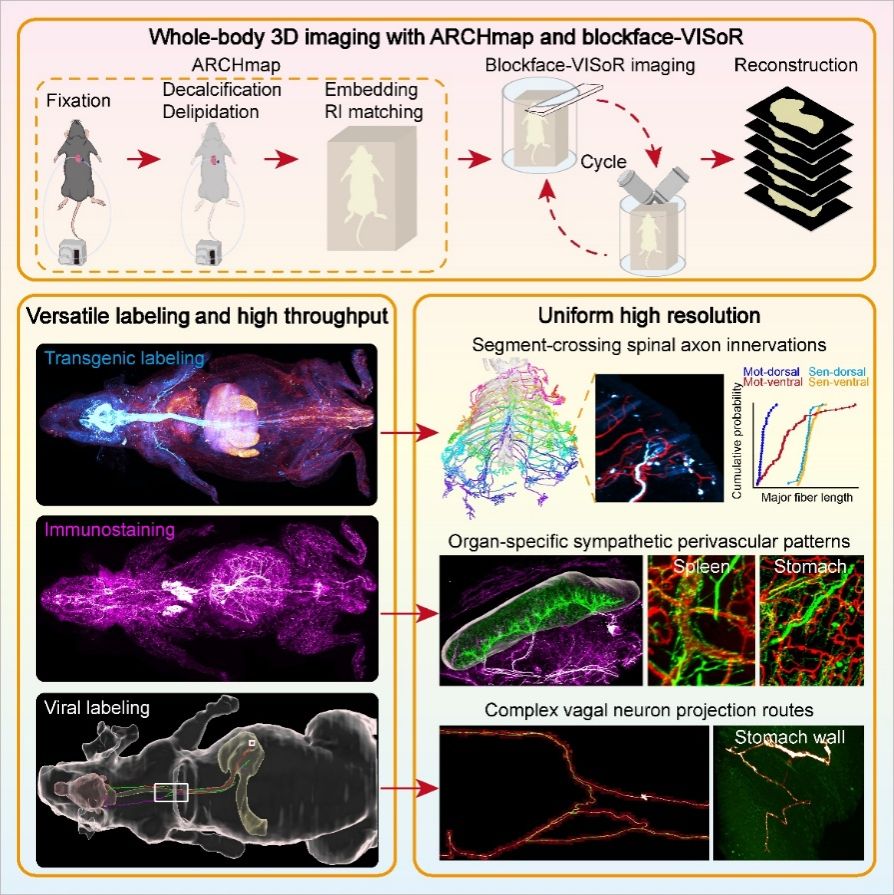
Whole-body Clearing and Blockface-VISoR Imaging Pipeline and Mesoscopic Connectomic Mapping of PNS
The researchers explained that this breakthrough technology not only helps establish a new paradigm for connectivity mapping of PNS and resolve fundamental questions in neural regulation, but also provide valuable insights into broader areas such as developmental biology, comparative anatomy, and biomedical research in general. Furthermore, there is still room for improvement and optimization of this technology. The next steps involve using two or more cameras for efficient multi-channel imaging, and exploring its application in imaging other larger-scale biological samples.
Cell journal reviewers praised: "These analyses produced strikingly detailed data at both the population and single-cell levels. Importantly, new insights emerged from this initial exploration," and "This is an interesting work, beautifully illustrated, and the methodology shows great potential."
Paper Link: https://doi.org/10.1016/j.cell.2025.06.011
Link to experimental techniques and imaging datasets: https://mesoanatomy.org/mesomouse/
(Editted by ZHAO Zheqian, English news center)